| Technical Name | Accurate multilocus sequence typing using Oxford Nanopore MinION with dual-barcode approach to multiplex large numbers of samples | ||
|---|---|---|---|
| Project Operator | National Health Research Institutes | ||
| Project Host | 廖玉潔 | ||
| Summary | We simultaneously sequenced seven full-length housekeeping genes of 96 methicillin-resistant Staphylococcus aureus (MRSA) isolates with dual-barcode multiplexing using just a single flow cell of Oxford Nanopore MinION, and then performed bioinformatic analysis (nanoMLST) for strain typing. The allele assignments obtained by nanoMLST were identical to those obtained by Sanger sequencing (359/359, 100%). It can be expected that nanoMLST will improve the study of epidemiology. |
||
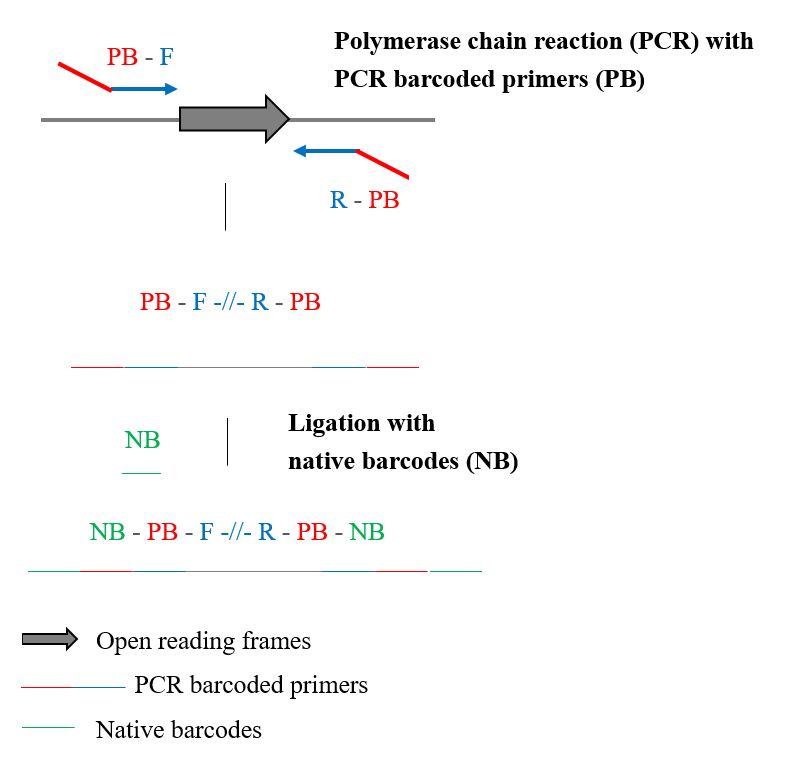
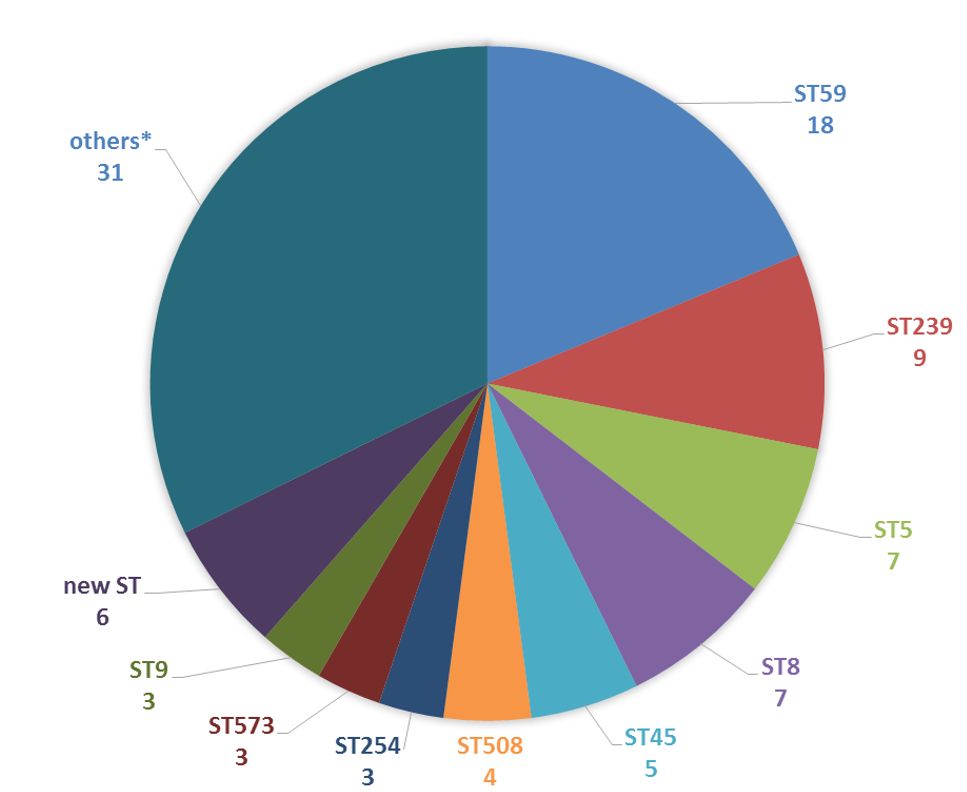
| Scientific Breakthrough | nanoMLST simultaneously types 96 methicillin-resistant Staphylococcus aureus isolates with dual-barcode using one single flow cell of ONT MinION. A total of 672 full-length genes have been sequenced with an average length of 1,300 bp and 100% typing accuracy could provide extra phylogenetic information for sub-typing. To our knowledge, this is the first study to perform high-throughput MLST accurately using MinION. |
||
| Industrial Applicability | nanoMLST is a new technique designed for MLST. It was successfully validated its typing accuracy on MLST of MRSA (100%). By exchanging the housekeeping genes and their matching databases to nanoMLST, one can easily characterize the sequence types of over 100 microbial species in just a few clicks after sequencing. The application could add a great value in hospital infection control measures, aquaculture, food industries and public health, etc. |
||
| Keyword | nanoMLST multilocus sequence typing (MLST) Oxford Nanopore Technologies (ONT) MinION next-generation sequencing (NGS) methicillin-resistant Staphylococcus aureus (MRSA) high-throughput targeted sequencing dual-barcode long read cost-effective | ||
- hanjie@nhri.org.tw
other people also saw