| Technical Name | High-Performance Deep Learning Pipeline Predicts Individuals in Mixtures of DNA Using Sequencing Data | ||
|---|---|---|---|
| Project Operator | National Taiwan University Centers of GenomicPrecision Medicine | ||
| Project Host | 李財坤 | ||
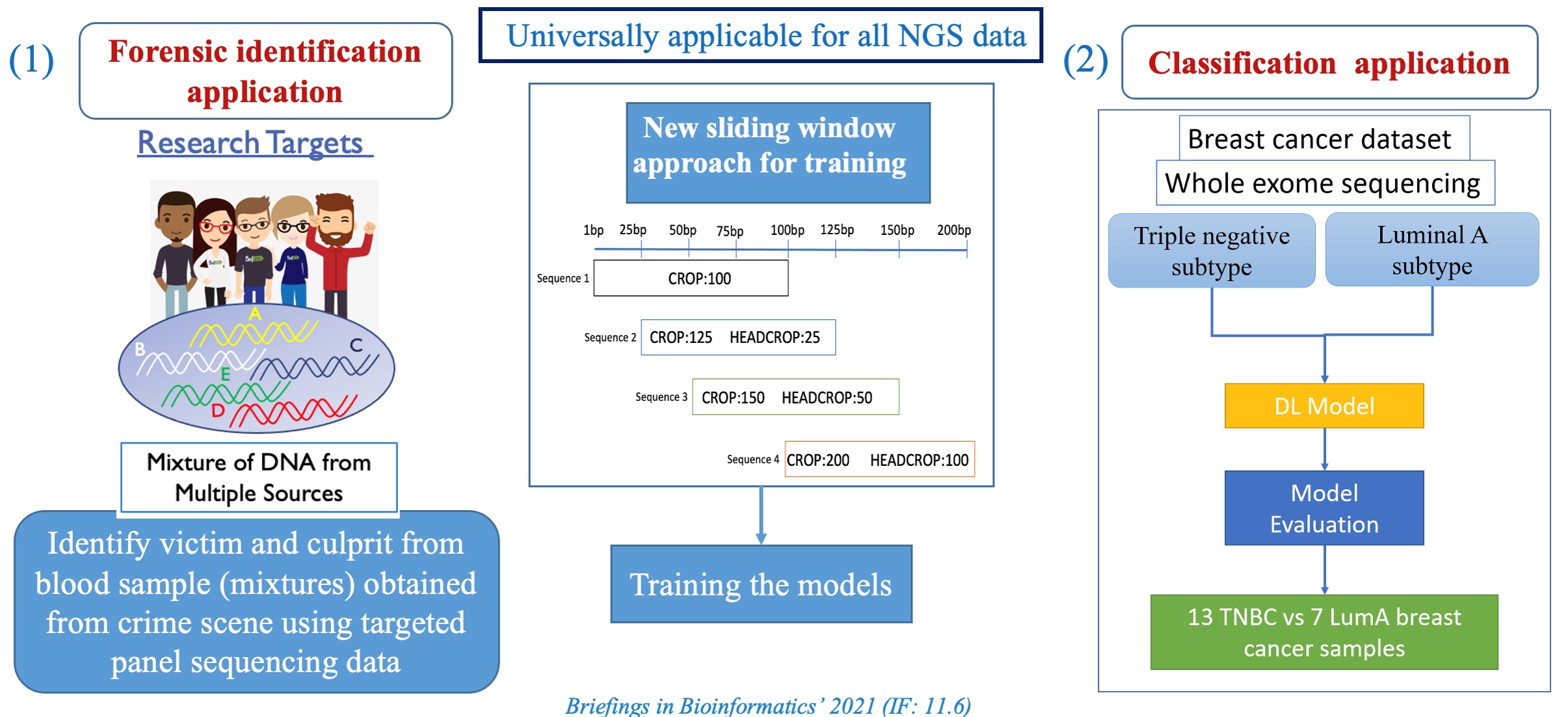
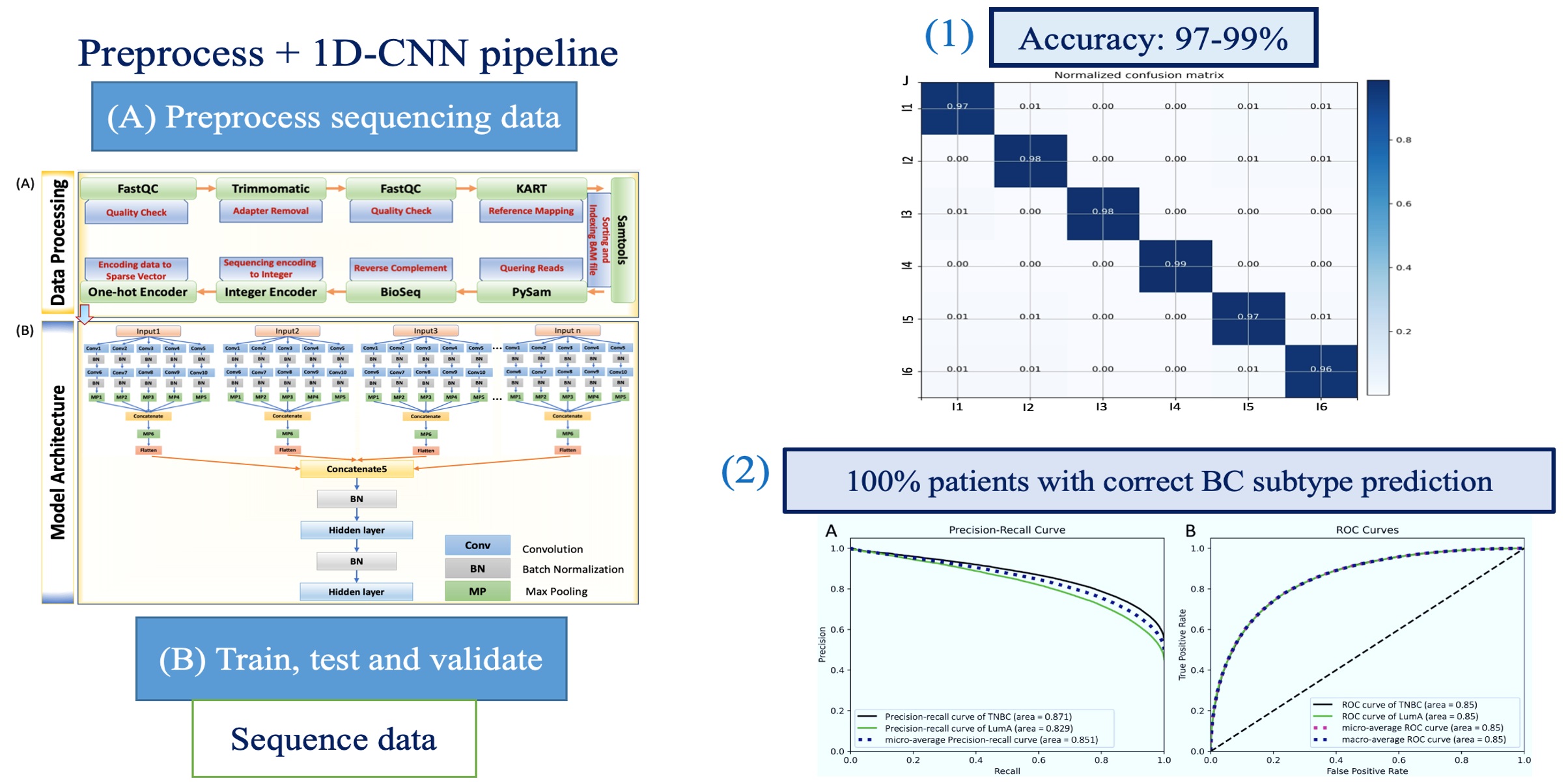
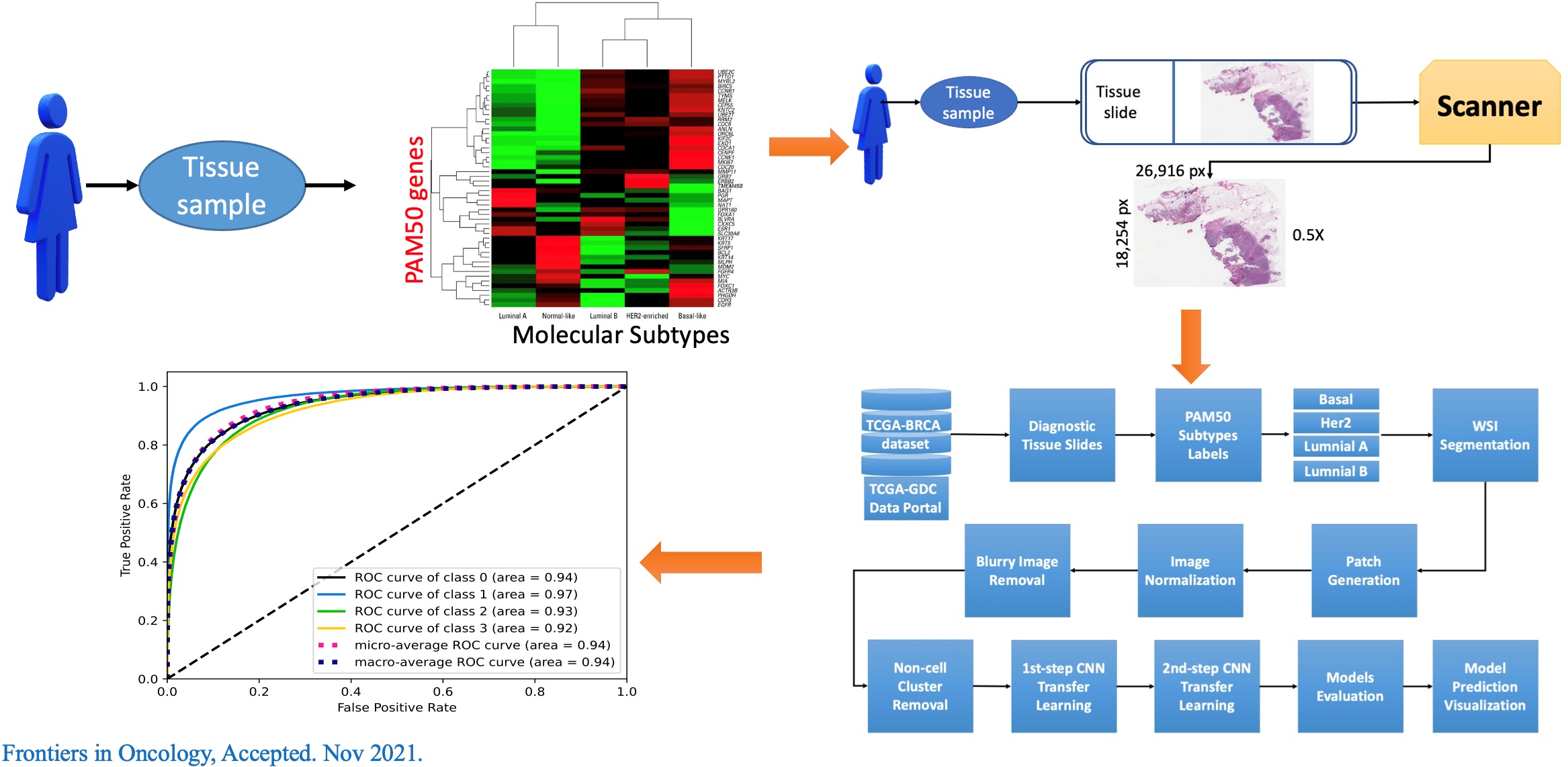
| Summary | We describe a 1-dimensional deep convolutional neural network (DCNN) that can potentially be applied globally to NGS data for the classification of individuals. The complete pipeline includes both sequencing data processing stepsdeep learning steps. To demonstrate the versatility of our proposed pipeline, we first applied our method on a forensic dataset, to identify individuals from highly imbalanced mixture of samples. Second, we used whole exome sequencing data from breast cancer patients to classify patients into triple negativeluminal A subtypes. To the best of our knowledge, this is one of the first attempts to use NGS read data with AI for classifyingdetecting majorminor contributors in mixed DNA samples. |
||
| Scientific Breakthrough | Our complete pipeline is applicable across different NGS platforms with state- of-the-art accuracy. Furthermore, a critical issue in training a deep learning model is the the sequence length input to the DL model should be identical for every sequence reads so that the number of features generated equally. To overcome this problem: (a) We introduced a sliding window approach to gener- ate identical sequence length fragments from original sequence reads (b) A customed sequence reads generator to combine these fragments into a single array for model input. This approach demonstrates that it improves the model performance dramatically. |
||
| Industrial Applicability | One of the potential applications of the current pipeline is highlighted in circulating tumor DNA detection in cell-free DNA samples which share the concept with our present study. Taking advantage of cfDNA sample only provides many benefits to the VAF methods such as no need of paired white blood cells sample as the background. This first reduce the cost for ctDNA estimation. Secondly, it helps to remove multiple steps of variant calling protocol that need white blood cells paired samples. In addition, taking advantage of learning from the sequence reads, the model can detect the tumor DNA at a very small quantity in comparison to variant calling methods, which is highly required the high coverage level from the samples. |
||
| Keyword | Gene sliding window convolutional neural network NGS sequencing data classification forensic | ||
- Contact
- Yee-Shin Lee
- yslee@ntu.edu.tw
other people also saw