| Technical Name | Hunt for Novel Antibiotics: Discover and design new anti-microbial peptides in AI | ||
|---|---|---|---|
| Project Operator | Academia Sinica | ||
| Project Host | 林仲彥 | ||
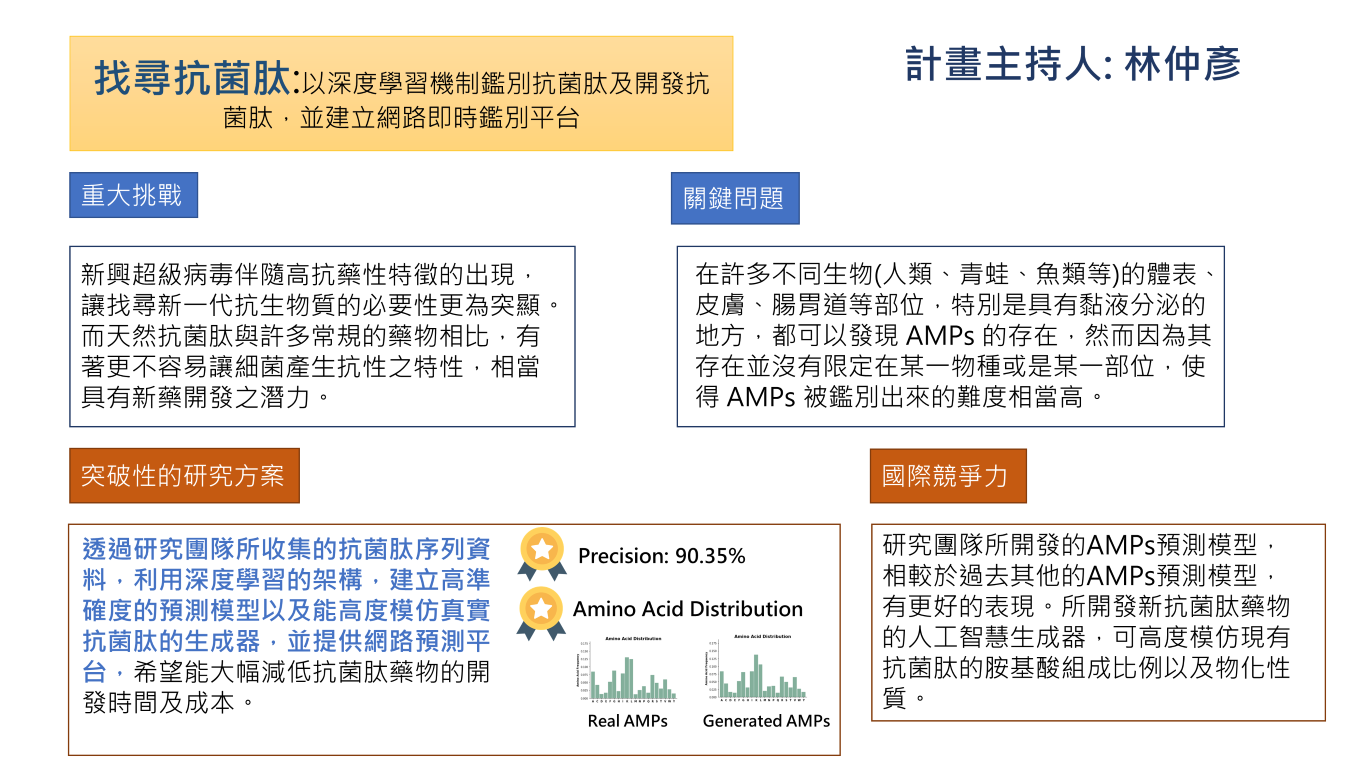
| Summary | 1.Antimicrobial peptides (AMPs) are innate immune components existing in many different organisms. They are promising drug candidates that can overcome drug resistance problems that pose threat to public health. However, identifying AMPs is difficult. To accelerate the process of discovering AMPs, we developed the following technology. |
||
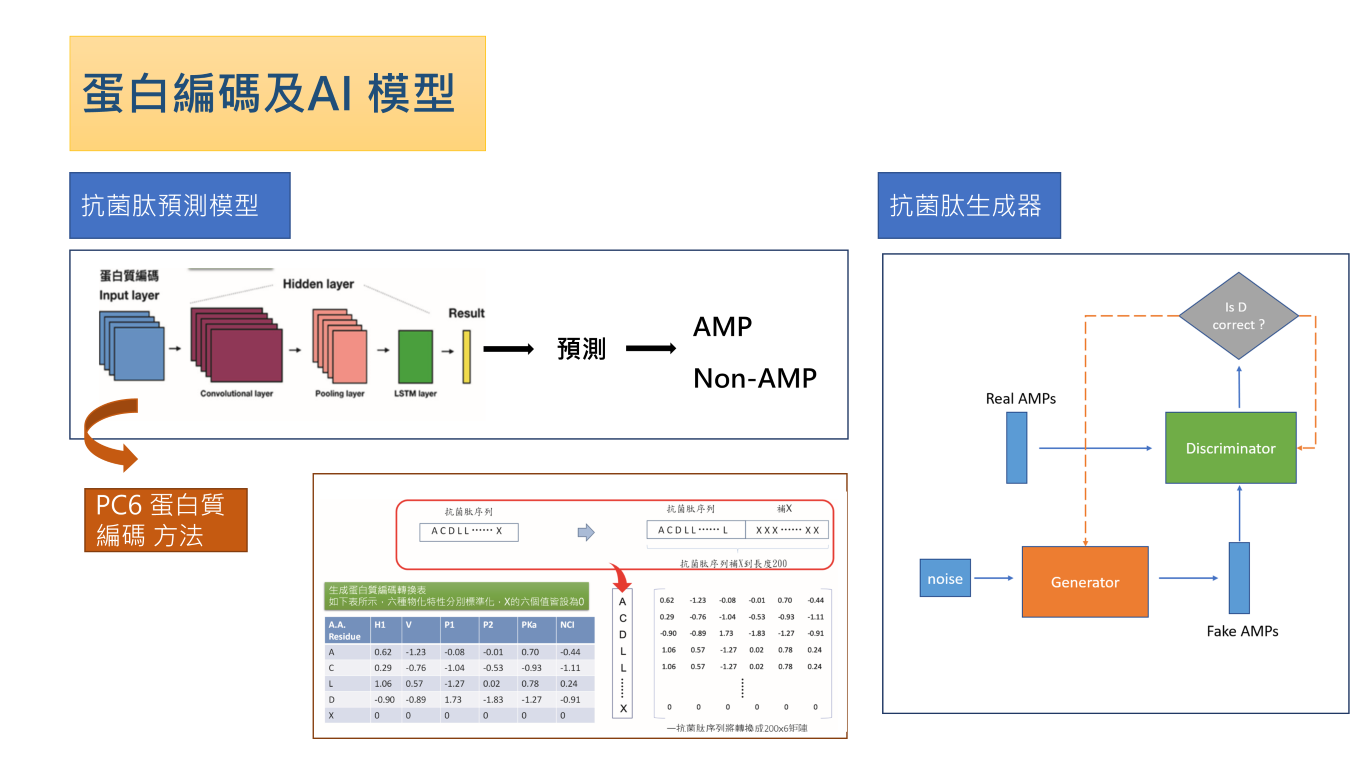
| Scientific Breakthrough | 1.We developed a new protein encoding method, PhysicoChemical Property (PC6), which considers both the order and the physicochemical properties of amino acids. This method improves the performance of machine learning. |
||
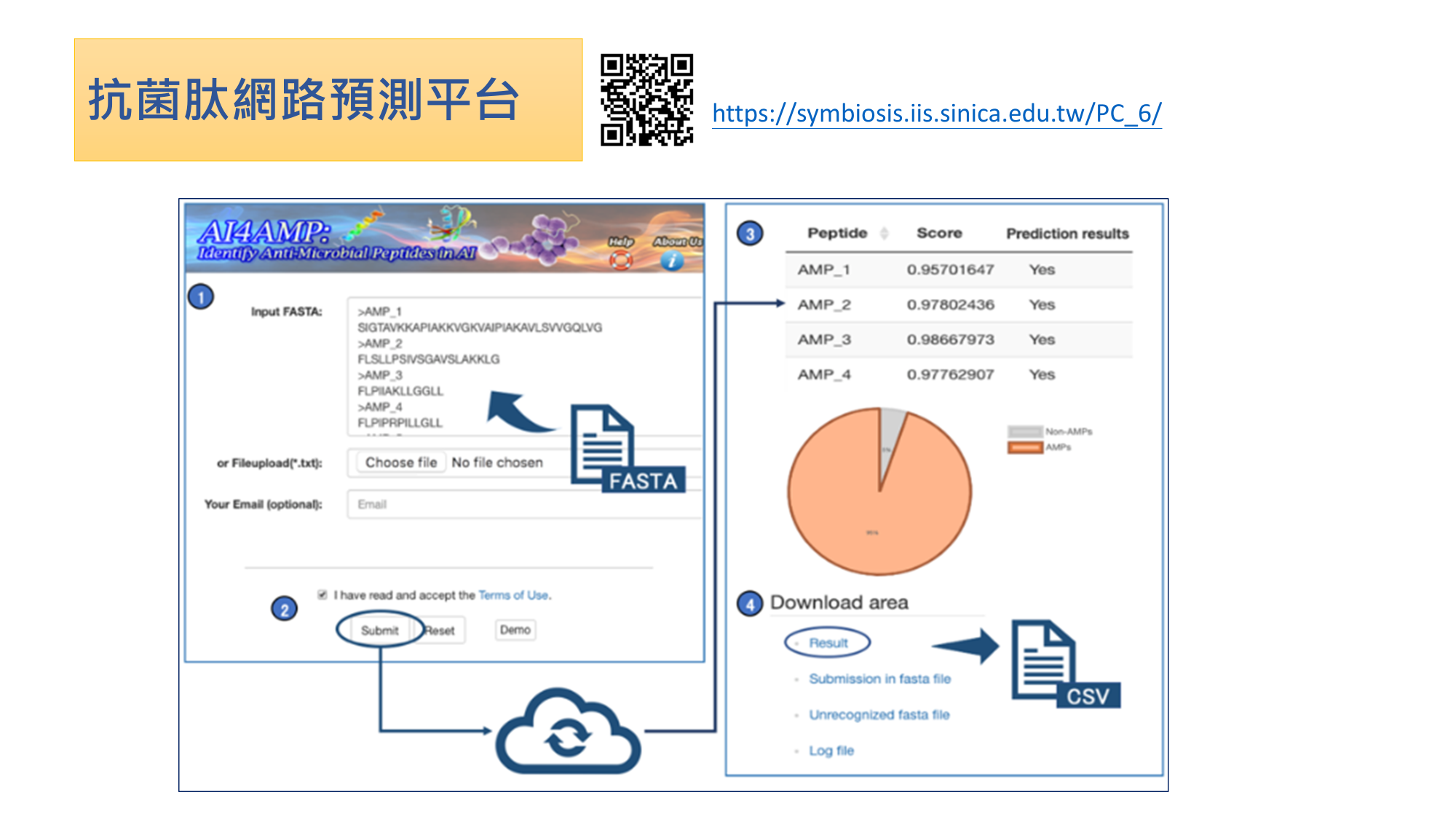
| Industrial Applicability | 1.Market potential and economic benefits: By combining the AMPs prediction platform and the antibacterial peptide generator, pharmaceutical factories and biotechnology companies will be able to obtain candidate protein sequences quickly and predict their antibacterial activities. This can greatly reduce the time and expense for antimicrobial drug development. |
||
| Keyword | Anti-Microbial Peptide deep learning PhysicoChemical Property Drug discovery generative adversarial network (GAN) peptide generator Novel protein sequence sequence encoding Superbugs novel antibiotics | ||
- cylin@iis.sinica.edu.tw
other people also saw